BioCompute Objects (BCO) is a community-driven project backed by the FDA (US Food and Drug Administration) and George Washington University to standardize exchange of High-Throughput-Sequencing workflows for regulatory submissions between FDA, pharma, bioinformatics platform providers and researchers.
 Members of the Research Object team (Carole Goble, Stian Soiland-Reyes, Michael R Crusoe) have been collaborating closely with the rest of the BCO community since 2016, in particular covering the integration of BCO with existing standards like Research Object, W3C PROV and Common Workflow Language.
Vahan Simonyan from FDA presented BioCompute Objects to the GA4GH Cloud workstream webinar on 2017-11-27.
The below blog post is based on an extract of Vahan’s slides modulated to cover my thoughts on the role of Research Objects with BCOs.
Members of the Research Object team (Carole Goble, Stian Soiland-Reyes, Michael R Crusoe) have been collaborating closely with the rest of the BCO community since 2016, in particular covering the integration of BCO with existing standards like Research Object, W3C PROV and Common Workflow Language.
Vahan Simonyan from FDA presented BioCompute Objects to the GA4GH Cloud workstream webinar on 2017-11-27.
The below blog post is based on an extract of Vahan’s slides modulated to cover my thoughts on the role of Research Objects with BCOs.
-
Introduction
-
Model of BioCompute Objects
-
JSON of BioCompute Object
-
Links and references
-
Packaging it up: Research Object
-
Take part!
Introduction
There is a particular challenge for regulatory bodies like FDA in areas like personalized medicine, as to review and approve the bioinformatics side they need to inspect and in some cases replicate the computational analytical workflow. The challenge here is not just the normal reproducibility thing about packaging software and providing required datasets, but also for human understanding of what has been done, by expressing the higher level steps of the workflow, their parameter spaces and algorithm settings. (PMC5510742)
Model of BioCompute Objects
At the heart of the BCO is a domain-specific object model which capture this essential information without going in details of the actual execution:
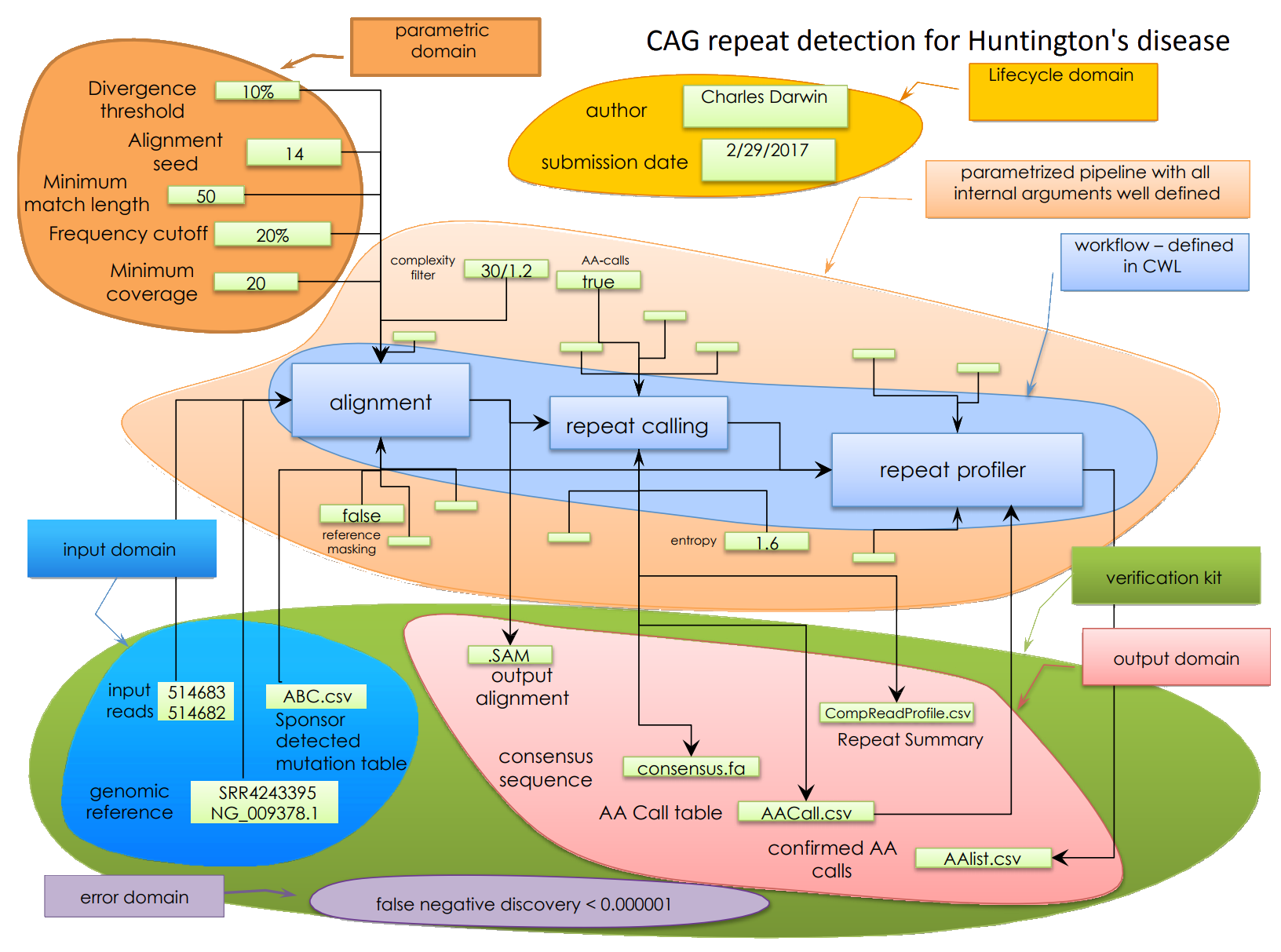 The full background and development of the BioCompute Object model is detailed in the recent BCO community paper (https://doi.org/10.1101/191783).
The full background and development of the BioCompute Object model is detailed in the recent BCO community paper (https://doi.org/10.1101/191783).
JSON of BioCompute Object
The BCO is expressed as a JSON format, which also includes additional metadata and external identifiers.
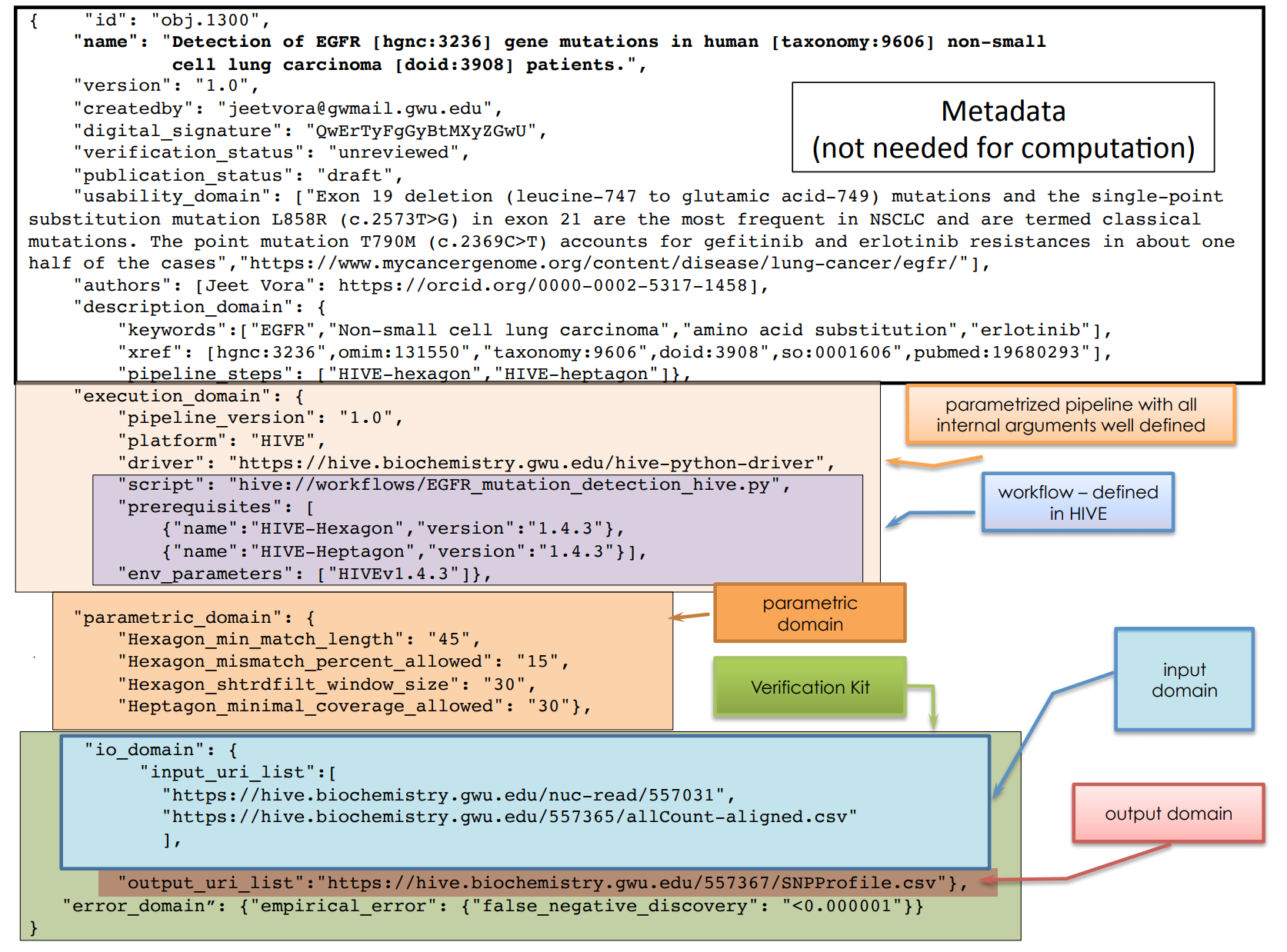 If we look at this JSON briefly, it is split into metadata, a brief overview of the pipeline with arguments and scripts. The actual workflow definition is defined outside. In addition we define parametric domain, and for verification the input/output domains. This allows inspection to see what is the scope of the analysis. For more details, see the first release of the BCO Specification Document (https://osf.io/zm97b/).
If we look at this JSON briefly, it is split into metadata, a brief overview of the pipeline with arguments and scripts. The actual workflow definition is defined outside. In addition we define parametric domain, and for verification the input/output domains. This allows inspection to see what is the scope of the analysis. For more details, see the first release of the BCO Specification Document (https://osf.io/zm97b/).
Links and references
Looking deeper, many of the BioCompute entries are actually external links or references: 

Packaging it up: Research Object
So we’ve identified that there are multiple references, attributions, provenance traces and annotations to keep track of in a BioCompute Object.
There is also the question of how to move the BCO around – the JSON has many external references as well as relative references to plain files – how can you capture it all without understanding all of the BCO spec?
We are looking at using the BagIt Research Objects for this purpose.
Bag-It is a digital archive format used by Library of Congress and digital repositories. It handles checksums and completeness of files, even if they are large or external.
Research Object (RO) (https://doi.org/10.1016/j.websem.2015.01.003) is a model for capturing and describing research outputs; embedding data, executables, publications, metadata, provenance and annotations. Although it is a general model, ROs have been used in particular for capturing reproducible workflows.
The combination of these, ro-bagit aka BDBags has recently been used by the NIH-BD2K-funded BDDS project for transferring and archiving very large HTS datasets (https://doi.org/10.1109/BigData.2016.7840618 [preprint]) in a location-independent way (the BDDS group has recently been awarded new funding under NIH Data Commons), so we hope BDBags could be a good choice also to to package and archive BCOs.
The below figure illustrates how the manifest of the Research Object (manifest.json) ties the BioCompute Object together:


Take part!
Do you want to take part in shaping Biocompute Objects? See the BioCompute Project Space at Open Science Framework, or join our community WebEx meetings (for call details, contact Charles Hadley King at: hadley_king@gwu.edu)